GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
4.6 (707) In stock
4.6 (707) In stock
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE
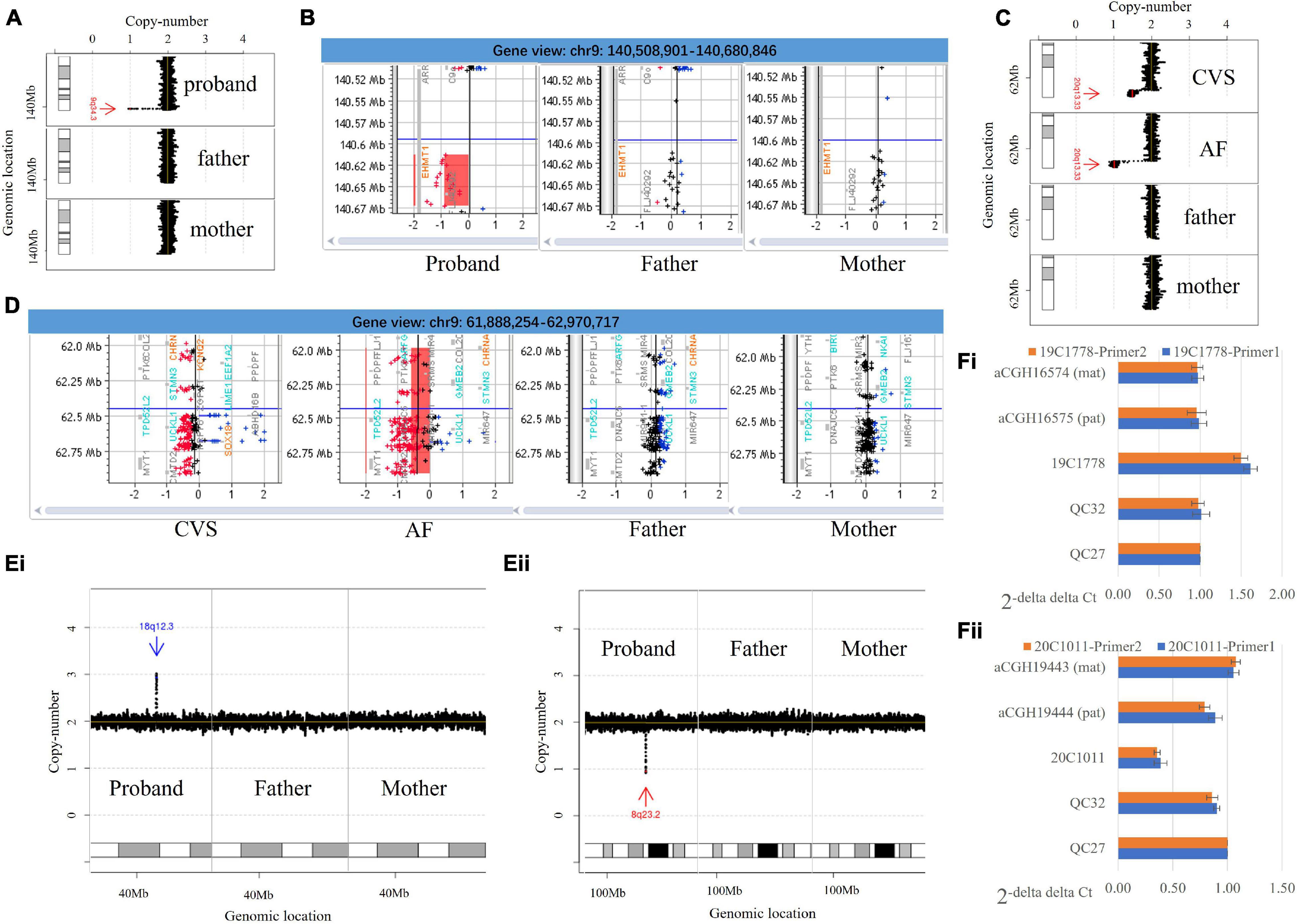
Frontiers Trio-Based Low-Pass Genome Sequencing Reveals Characteristics and Significance of Rare Copy Number Variants in Prenatal Diagnosis

Ensemble of nucleic acid absolute quantitation modules for copy number variation detection and RNA profiling
PDF) CNHplus: the chromosomal copy number heterogeneity which respects biological constraints
PDF) WAVECNV: A New Approach for Detecting Copy Number Variation by Wavelet Clustering

PDF) WAVECNV: A New Approach for Detecting Copy Number Variation by Wavelet Clustering
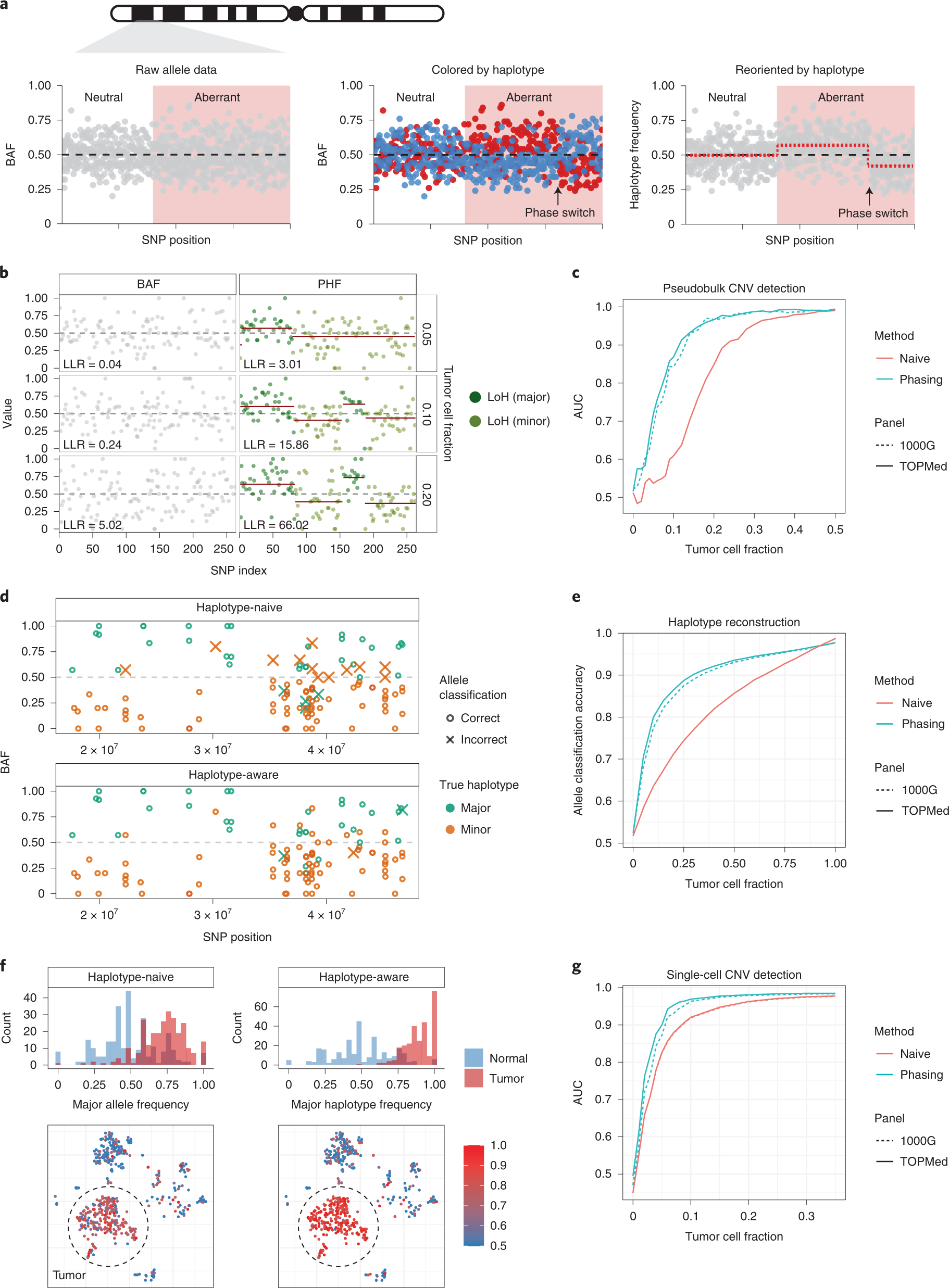
Haplotype-aware analysis of somatic copy number variations from single-cell transcriptomes

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data

Comprehensive Assessment of Somatic Copy Number Variation Calling Using Next-Generation Sequencing Data

Copy number analysis by low coverage whole genome sequencing using ultra low-input DNA from formalin-fixed paraffin embedded tumor tissue, Genome Medicine

PDF) HBOS-CNV: A New Approach to Detect Copy Number Variations From Next-Generation Sequencing Data
QIAGEN Omicsoft Copy Number Variation Analysis tutorial

PDF) HBOS-CNV: A New Approach to Detect Copy Number Variations From Next-Generation Sequencing Data

American Society for Clinical Pharmacology and Therapeutics - 2019 - Clinical Pharmacology & Therapeutics - Wiley Online Library

Copy-number variants in clinical genome sequencing: deployment and interpretation for rare and undiagnosed disease